SINEBase is a database of short interspersed elements and tools for their identification and analysis. It is intended to:
SINEBase is not intended to:
Retro(trans)posons are genetic elements that can amplify themselves in eukaryotic genomes via an RNA intermediate, which requires their transcription and reverse transcription. Retroposons are divided into three classes: LTR elements, LINEs, and SINEs. The elements that encode reverse transcriptase (RT), an enzyme providing for the reverse transcription and integration of the DNA copy into the genome, are called autonomous transposons. Nonautonomous retroposons rely on the RTs of autonomous transposons. LTR transposons and LINEs can be autonomous or nonautonomous; and their genomic copies are transcribed by the cellular RNA polymerase II.
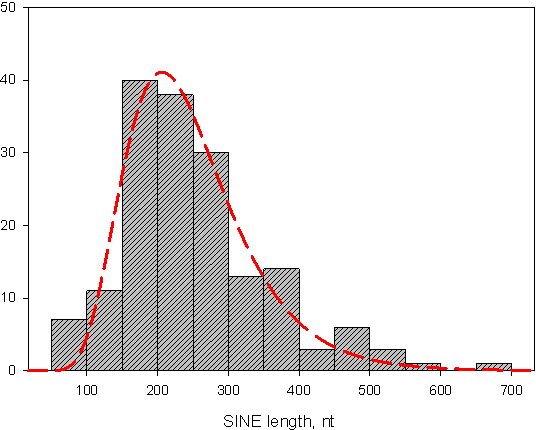
Short interspersed elements (SINEs) are defined as relatively short (< 700 bp) nonautonomous retroposons transcribed by the cellular RNA polymerase III (pol III) from an internal promoter, while their reverse transcription depends on the RT of partner LINEs. Eukaryotic genomes can harbor hundreds thousands (sometimes more) of SINE copies; copies originating from a common ancestral SINE can differ from each other by single-nucleotide alterations as well as by longer internal deletions or duplications (SINEs with such duplication are called quasidimeric). Some of them can become founders of new SINE subfamilies.
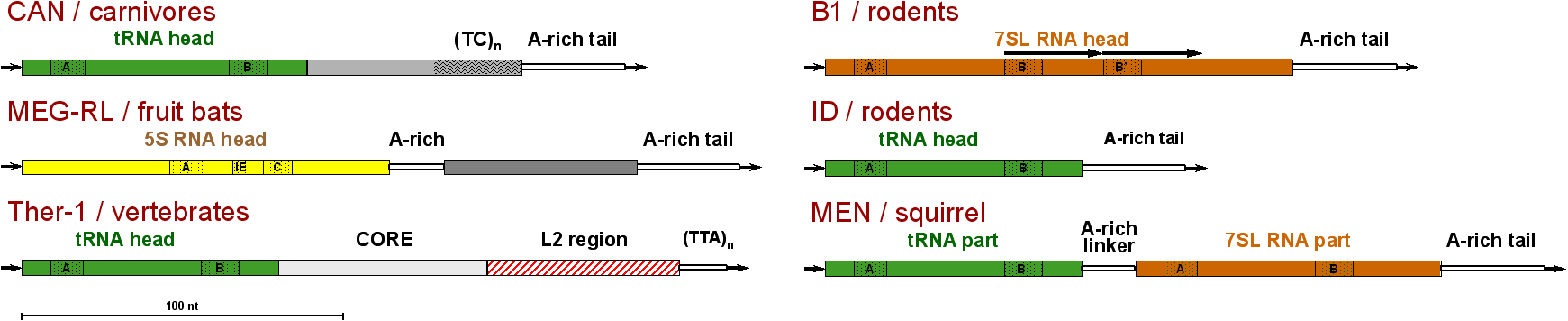
SINEs consist of two or more modules; typically, head, body, and tail. The 5'-terminal head originates from the cellular RNAs synthesized by pol III: tRNA, 7SL RNA, or 5S rRNA. The origin of the body is either unknown or it descends from a partner LINE. SINEs with a LINE-derived region mimic LINE RNA in the reverse transcription (such SINEs belong to the stringent group). It can also contain a domain shared by distant SINE families (CORE and similar domains). The 3'-terminal tail is a sequence of variable length consisting of simple (often degenerate) repeats. In addition, two SINEs can combine into a dimeric SINE, thus, giving rise to a new SINE family. SINEs consisting of the head and tail only are called simple, while dimeric, trimeric, etc. are complex SINEs.
SINEs should be distinguished from RNA pseudogenes: the pseudogenes are generated by the reverse transcription of the cellular RNAs (e.g., 5S rRNA) rather than of SINE RNAs transcribed from their genomic copies. In practical terms, most SINEs have extra (body) sequences, while simple SINEs have characteristic substitutions/indels shared with their source gene but not with the cellular RNA gene. In addition, SINEs significantly outnumber RNA pseudogenes.
The notion of ‘SINE family’ is widely used but not clearly defined. We consider SINE family as a set of SINEs
Thus, similar SINEs with different LINE-derived regions belong to different families. Long insertions are considered as modules. At the same time, internal deletions or duplications within modules do not give birth to a new family; although a combination of complete or almost complete SINEs (complex SINEs) is considered as a new family (thus, pB1 and quasidimeric B1 are subfamilies of the same family, while dimeric Alu represents a distinct family). Finally, there are а few SINEs with quite similar structure but of independent origin (certain simple SINEs), which are considered as different families.
Database of SINE families is presented as a Table integrating the following data:
The Table contents can be filtered and sorted in many ways. E.g., you can limit it to mammalian SINE families with a LINE-derived region and sort them by length (see Tips & Tricks)
Note that SINEs of the same family have the same modules in the same order; at the same time, they can have relatively small deletions or internal duplications. The tail length and even sequence is not a marker of SINE families.
SINESearch is a FASTA-based search tool that utilizes simple parameters to select sequences of interest instead of the internal FASTA's statistical significance test. This obviates two limitations of FASTA (as well as BLAST etc.) in the case of relatively short and degenerate similarities between nucleotide sequences of SINEs:
Finally, the parameters used, overlap length and sequence identity, are biologically sensible and allow easy adjustment of hit selection.
SINESearch is simple to use and fast. Specify the search parameters, bank to search, and query sequence, and press the '' button to start search. If error message appears, press the '' button, enter correct data ('' button can be used to reset all fields), and press the '' button again. The results are sorted by the best fit coefficient (reflecting correspondence between the total lengths of the sequences and the overlap length; note that it does not directly depend on the sequence identity). However, the results can be sorted by other parameters (sequence name, identity, or overlap) by clicking on column headers marked with ![]() . In the case of the SINEBase bank, the output contains links to the SINE Table, where you can find details about the SINE families found. If you are not satisfied with the results, try to adjust parameters or redefine the query sequence limits.
. In the case of the SINEBase bank, the output contains links to the SINE Table, where you can find details about the SINE families found. If you are not satisfied with the results, try to adjust parameters or redefine the query sequence limits.
SINESearch input fields:
Sequence identity. Allowed range: 40-100%. Generally, it is not recommended to decrease this value below 65%. The default value is 65% but it automatically changes to 60% for the RNA banks.
Min overlap lenght. Allowed range: 30-1000 nt. Use 90% of the query sequence length as the starting point; 60 nt is recommended for the RNABase bank; use common sense: some modules can be as short as ~30 nt, while others are longer. The default value is 70 nt but it automatically decreases to 60 and 40 nt for the RNA and CORE banks, respectively.
Sequence Entry. Query sequence can be entered manually (typed or pasted) or uploaded from a local file. The sequence must be in FASTA format. Only IUPAC nucleotide symbols are allowed. The maximum sequence length is 2 kb. Multiple query sequences are not supported. Either enter or upload query sequence; when a sequence file is uploaded the manual sequence entry field is disabled and vice versa (use the '' buttons to reset the file upload box or the manual sequence entry fields; while '' resets all fields). The query sequence can be shortened from both ends using the Offset parameters (notice that the numbering of the full-length sequence is preserved).
TSDSearch is a tool to search for relatively short target site duplications of genomic DNA that commonly frame retrotransposons including SINEs. Found repeats are shown as arrows below the sequence ruler and as sequences with coordinates. TSDs are sorted by a compromise between total length and length of matches, so that 'best' TSDs are list first.
Technically, TSDSearch is implemented in JavaScript, which means that the calculation is performed by your web browser/computer. Clearly, JavaScript should be enabled in your browser. Finally, execution time significantly varies bewteen different browsers. Overall, more recent browsers execute TSDSearch faster. Chrome showed the best performance among popular browsers tested.
Search Area. A typical task in SINE analysis is to identify TSDs framing SINE sequence(s). In this context, the region where TSDs are searched should not include the proper SINE sequence as well as areas too distant from it. Blind analysis of the whole region not only substantially increases the computation time but also complicates data intepretation. Setting the 5' and 3' offset values as well as the lengths of regions to analyze (ranges) makes it possible to focus on the desired areas. Bear in mind that the tails can substantially vary in length, so it is a good practice to increase the 3' range relative to the 5' range. On the other hand, the 5' range has the greatest impact on calculation time so do not increase this value unless nessesary.
TSD parameters. The search algorithm considers TSD as three blocks of nucleotides (subrepeats) identical between the 3' and 5' TSDs (i.e., subrepeat 1 in 5' TSD is identical to subrepeat 1 in 3' TSD etc.). Subrepeats can be separated by variable spacers. For instance, 5' TSD: (ACCT)a(GGG)(TAC) and 3' TSD: (ACCT)(GGG)ac(TAC); subrepeats are shown in parentheses and spacers are in lowercase. Subrepeats cannot be shorter that the subrepeat min lengths specified, while spacers cannot be longer than the max length of spacers. Min length of total match (without gaps) and max total mismatch length allow fine tunung of TSD length and similarity, respectively. Finally, the number of displayed TSDs for a query sequence is limited by the max number of TSDs (specify '1' to show the best TSD only).
Sequence Entry. Query sequence can be entered manually (typed or pasted) or uploaded from a local file (if you use a recent browser that supports HTML5 file operations). The sequence must be in FASTA format. Only A, C, G, and T nucleotide symbols are allowed in the search area. Notice that U, N, and X are not allowed. Gaps ('~', '–', & ' ') are allowed and ignored. Multiple sequence query can be analyzed, but all sequences must have unique names. For good reason, all sequences should be longer than the sum of the left and right offsets and ranges. Either enter or upload query sequence; when a sequence file is uploaded the manual sequence entry field is cleared and vice versa (use the '' buttons to reset the file upload box or the manual sequence entry fields; while '' resets all fields).
All parameters and sequences are checked prior to TSD search. Analysis will not start if any parameter or sequence does not conform to the requirements. In this case, balloon message(s) appear near the field to be corrected.
We encourage the submission of new data on SINE families. Please, make sure that your SINE comply with the requirements and provide all nessesary information, which includes submitter's data (name, affiliation etc.), SINE data, and publication (if any). SINE data includes the SINE family name, consensus sequence, taxonomic distribution, copy number, tail repeat unit, and comments.
As you proceed to the next field (as well as when you press the '' button), error prompts may appear (e.g., 'This field is required' or 'Invalid email address'). You won't be able to send data without fulfilling all the requirements. Please, contact us if you find some requirements irrelevant (e.g., you have discovered a 55-nt SINE). If no (more) errors, pressing the '' button sends your data to the SINEBase. You will promptly recieve an automatic confirmation e-mail. Please, allow some time for us to review your submission.